nn-UNet
Overview
nn-UNet is a 2d/3d U-NET library designed to segment medical images, refer to github and the following citation:
Isensee, F., Jaeger, P. F., Kohl, S. A., Petersen, J., & Maier-Hein, K. H. (2021). nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods, 18(2), 203-211.
nn-UNet is easiest to use with their command line interface with three commands nnUNetv2_plan_and_preprocess,
nnUNetv2_train and nnUNetv2_predict.
For cvpl_tools, cvpl_tools/nnunet/cli.py provides two
wrapper command line interface commands train and predict that simplify the three commands into
two and hides unused parameters for SPIMquant workflow.
cvpl_tools/nnunet needs torch library and pip install nnunetv2. GPU is automatically used when
nnUNetv2_train and nnUNetv2_predict are called directly or indirectly through train and
predict and when you have a GPU available on the computer.
For those unfamiliar, nn-UNet has the following quirks:
Residual encoder is available for nnunetv2 but we prefer without it since it costs more to train
Due to limited training data, 2d instead of 3d_fullres mode is used in
cvpl_toolsIt trains on images pairs of input size (C, Y, X) and output size (Y, X) where C is number of color channels (1 in our case), and Y, X are spatial coordinates; specifically, N pairs of images will be provided as training set and a 80%-20% split will be done for train-validation split which is automatically done by nnUNet. It should be noted in our case we draw Z images from a single scan volume (C, Z, Y, X), so a random split will have training set distribution correlated with validation set generated by nnUNet, but such thing is hard to avoid
The algorithm is not scale-invariant, meaning during prediction, if we zoom the input image by a factor of 2x or 0.5x we get much worse output results. For best results, use the same input/output image sizes as the training phase. In our mousebrain lightsheet dataset, we downsample the original >200GB dataset by a factor of (4, 8, 8) before running the nnUNet for training or prediction.
The algorithm supports the following epochs, useful for small-scale training in our case: link if you input number of epochs not listed in this page to the
predictcommand, an error will occurnn-UNet supports 5-fold ensemble, which is to run
nnUNetv2_traincommand 5 times each on a different 80%-20% split to obtain 5 models to ensemble the prediction. This does not require rerunnnUNetv2_plan_and_preprocessand is supported by the--foldargument ofcvpl_tools’traincommand so you don’t need to run it 5 times. If you finish training all folds, you may use the--foldargument ofcvpl_tools’predictcommand to specifyallfor better accuracy after ensemble or0to specify using the first fold trained for comparison.Running the nn-UNet’s command
nnUNetv2_trainorcvpl_tools’traingenerates onennUNet_resultsfolder, which contains a model (of size a few hundred MBs) and a folder of results including a loss/DICE graph and a log file containing training losses per epoch and per class. The same model file is used later for prediction.
Negative Masking for Mouse-brain Lightsheet
In this section, we focus primarily on the usage of nn-UNet within cvpl_tools. This part of the
library is designed with handling mouse-brain lightsheet scans in mind. These scans are large (>200GB)
volumes of scans in the format of 4d arrays of data type np.uint16 which is of shape (C, Z, Y, X). An
example is in the google storage bucket
“gcs://khanlab-lightsheet/data/mouse_appmaptapoe/bids/sub-F4A1Te3/micr/sub-F4A1Te3_sample-brain_acq-blaze4x_SPIM.ome.zarr”
with an image shape of (3, 1610, 9653, 9634).
The objective of our algorithm is to quantify the locations and sizes of beta-amyloid plaques in a volume of lightsheet scan like the above, which appear as small-sized round-shaped bright spots in the image volume, and can be detected using a simple thresholding method.
Problem comes, however, since the scanned mouse brain edges areas are as bright as the plaques, they will be marked as false positives. These edges are relatively easier to detect by a UNet algorithm, which results in the following segmentation workflow we use:
For N mousebrain scans M1, …, MN we have at hand, apply bias correction to smooth out within image brightness difference caused by imaging artifacts
Then select one of N scans, say M1
Downsample M1 and use a GUI to paint a binary mask, which contains 1 on regions of edges and 0 on plaques and elsewhere
Split the M1 volume and its binary mask annotation vertically to Z slices, and train an nnUNet model on these slices
Above produces a model that can predict negative masks on any mousebrain scans of the same format; for the rest N-1 mouse brains, they are down-sampled and we use this model to predict on them to obtain their corresponding negative masks
These masks are used to remove edge areas of the image before we apply thresholding to find plaque objects. Algorithmically, we compute M’ where
M'[z, y, x] = M[z, y, x] * (1 - NEG_MASK[z, y, x]) for each voxel location (z, y, x); then, we apply threshold on M’ and take connected component of value of 1 as individual plaque objects; their centroid locations and sizes (in number of voxels) are summarized in a numpy table and reported
In this next part, we discuss the annotation (part 2), training (part 3) and prediction (part 4).
Annotation
Data quality is the most crucial to accurate predictions when training supervised models, in which case this is relevant to us in terms of how well we can annotate 3d image volumes at hand. Our annotation is the negative masking of edge areas of the brain to remove edges before applying simple thresholding. We model how good an annotation of negative mask by looking at:
For the simple threshold of choice t, how many voxels are above the threshold across the entire image, say V
The number of voxels covered by plaques areas above threshold t, and how many of them are correctly annotated as 0, and how many of them are incorrectly annotated as 1
The number of voxels covered by brain edge areas above threshold t, and how many of them are correctly annotated as 1, and how many of them are incorrectly annotated as 0
these metrics are best summarized as IOU or DICE scores. A DICE score curve can be obtained in training process, automatically generated by nn-UNet. We look at an example segmentation below.
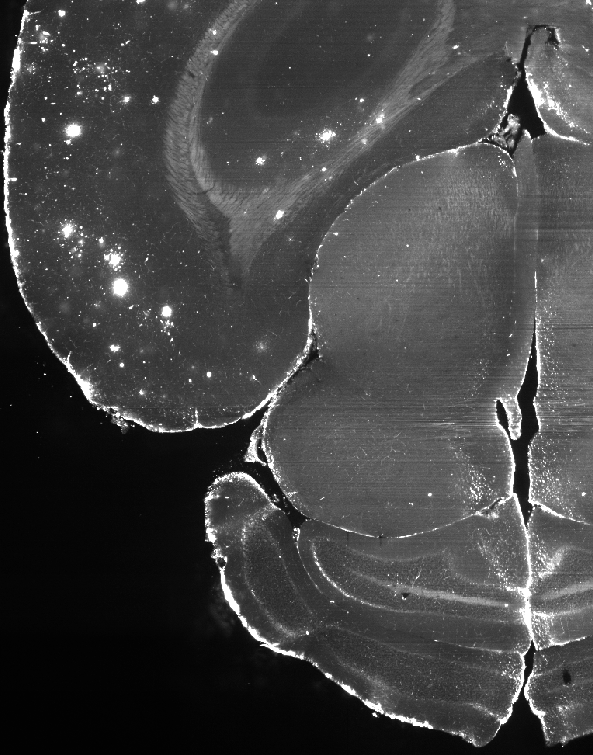
Slice of the mouse brain, not annotated (without negative masking)
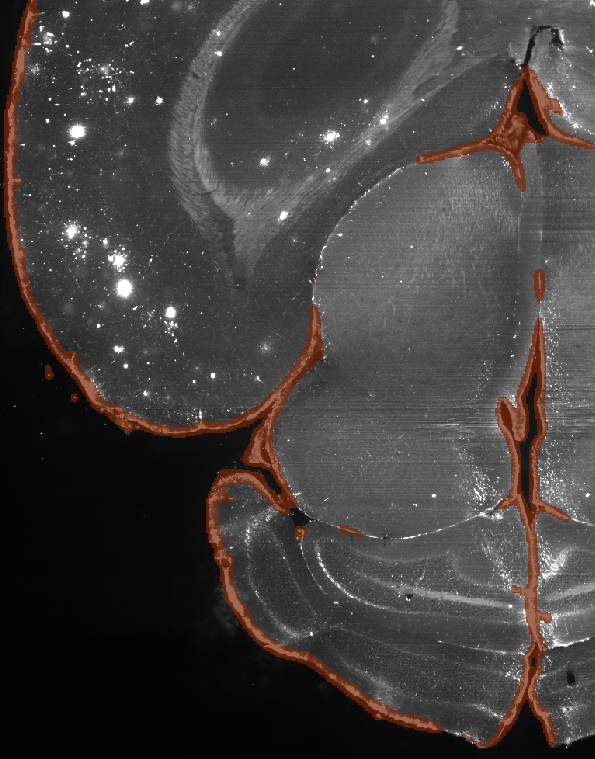
Slice of the mouse brain, annotated (with negative masking)
Here the algorithm, as intended, marks not only the outer edges of the brain but also some of the brighter inner structures as edge areas to be removed, since they can’t be plaques. The bright spots on the upper left of the images are left as is, for they are all plaques. Overall, the annotation requires quite a bit of labour and it is preferred to obtain a high quality annotated volume over many low quality ones.
In cvpl_tools, the annotation is done using a Napari based GUI with a 2d cross-sectional viewer and
ball-shaped paint brush. Follow the following steps to get started:
In a Python script, prepare an image you would like to annotate
im_annotatein Numpy array format, which may requires downsample the original image:
import cvpl_tools.nnunet.lightsheet_preprocess as lightsheet_preprocess
# original image is, say, an OME ZARR image of size (3, 1610, 9653, 9634)
OME_ZARR_PATH = 'gcs://khanlab-lightsheet/data/mouse_appmaptapoe/bids/sub-F4A1Te3/micr/sub-F4A1Te3_sample-brain_acq-blaze4x_SPIM.ome.zarr'
BA_CHANNEL = 0 # only the first channel is relevant to Beta-Amyloid detection
FIRST_DOWNSAMPLE_PATH = 'o22/first_downsample.ome.zarr' # path to be saved
first_downsample = lightsheet_preprocess.downsample(
OME_ZARR_PATH, reduce_fn=np.max, ndownsample_level=(1, 2, 2), ba_channel=BA_CHANNEL,
write_loc=FIRST_DOWNSAMPLE_PATH
)
print(f'Shape of image after downsampling: {first_downsample.shape}')
Ideally the downsampled image should also go through n4 bias correction before the next step.
Next, convert the image you just downsampled to a numpy array, and use
annotatefunction to add layers to a napari viewer and start annotation:
from cvpl_tools.nnunet.annotate import annotate
import cvpl_tools.ome_zarr.io as ome_io
import napari
viewer = napari.Viewer(ndisplay=2)
im_annotate = first_downsample.compute() # this is a numpy array, to be annotated
ndownsample_level = (1, 1, 1) # downsample by 2 ^ 1 on three axes
# image layer and canvas layer will be added here
annotate(viewer, im_annotate, 'o22/annotated.tiff', ndownsample_level)
viewer.show(block=True)
Note saving is manual, press ctrl+shift+s to save what’s annotated (which creates a tiff
file “o22/annotated.tiff”). im_annotate is lightsheet image first corrected by bias,
then downsampled by levels (1, 2, 2) i.e. a factor of (2, 4, 4) in three directions to a size
that can be conveniently displayed locally, in real-time and without latency.
In this example, we choose to use a binary annotation volume of shape (2, 2, 2) times smaller than the original image in all three directions. This is to save space during data transfer. Later nn-UNet will also need image of same shape as the annotation, so we also want to keep a further downsampled image file that is the same size as the annotation. We will see this in the training section below.
Due to the large image size, you may need multiple sessions in order to completely annotate one scan. This can be done by running the same code in step 2, which will automatically load the annotation back up, and you can overwrite the old tiff file with updated annotation by, again,
ctrl+shift+s
Training
In the above annotation phase, we obtained two dataset: one is the annotated tiff volume at path
'o22/annotated.tiff', the other is the downsampled image at path ‘o22/first_downsample.ome.zarr’. We
will use the latter as the training images and the former as the training labels for nn-UNet training.
Here the images need to be once further downsampled in order to match image and label volume shapes:
import cvpl_tools.nnunet.lightsheet_preprocess as lightsheet_preprocess
FIRST_DOWNSAMPLE_PATH = 'o22/first_downsample.ome.zarr' # path to be saved
SECOND_DOWNSAMPLE_PATH = 'o22/second_downsample.ome.zarr'
second_downsample = lightsheet_preprocess.downsample(
FIRST_DOWNSAMPLE_PATH, reduce_fn=np.max, ndownsample_level=(1, 1, 1), ba_channel=BA_CHANNEL,
write_loc=SECOND_DOWNSAMPLE_PATH
)
Next, we feed the images to nn-UNet for training. This requires torch installation and a GPU on the computer.
import cvpl_tools.nnunet.triplanar as triplanar
train_args = {
"cache_url": 'nnunet_trained', # this is the path to which training files and trained model will be saved
"train_im": SECOND_DOWNSAMPLE_PATH, # image
"train_seg": 'o22/annotated.tiff', # label
"nepoch": 250,
"stack_channels": 0,
"triplanar": False,
"dataset_id": 1,
"fold": '0',
"max_threshold": 7500.,
}
triplanar.train_triplanar(train_args)
250 epochs takes less than half a day to run on a consumer GPU.
Prediction
In the training phase we trained our model in the 'nnunet_trained' folder. In this folder not everything
is required for prediction, but only the model file in the path
nnunet_trained/train/yx/nnUNet_results/Dataset001_Lightsheet1/nnUNetTrainer_250epochs__nnUNetPlans__2d/fold_0/checkpoint_final.pth
is required. Therefore to reduce file size when you copy this file to other machines for inference, you can
remove the raw and preprocessed folder as well as the checkpoint_best.pth model. Pack the nnunet_trained
folder for prediction, as you will need to specify the this path during prediction.
nn-UNet prediction takes 3 main arguments:
Path to your nn-UNet trained folder
is the tiff file to predict
output tiff path
Below gives an example snippet carrying out the prediction on tiff images:
import cvpl_tools.nnunet.triplanar as triplanar
pred_args = {
"cache_url": 'nnunet_trained',
"test_im": SECOND_DOWNSAMPLE_CORR_PATH,
"test_seg": None,
"output": 'output.tiff',
"dataset_id": 1,
"fold": '0',
"triplanar": False,
"penalize_edge": False,
"weights": None,
"use_cache": False,
}
triplanar.predict_triplanar(pred_args)
Here we are predicting on the training set at SECOND_DOWNSAMPLE_CORR_PATH. In practice we replace this with other downsampled and corrected mousebrain lightsheet scan volumes. The prediction will automatically use CPU if GPU is not available; or use GPU if one is. Output tiff can be found at ‘output.tiff’, which should be the same size as input volume.
Tips on prediction quality:
1. Five fold training or prediction can be specified by setting “fold” to “all”. This will improve accuracy slightly but takes 5 times the computation resource to train or predict.
2. The tri-planar option will predict the volume in z/y/x three ways and merge the results, which takes 3 times the computation to train or predict. This significantly increases accuracy, but the result mask is often not desirable. This is because the ensembed mask often flickers in local areas and can affect contour counting in our application, and is harder to interpret when looking through yx cross-sectional plane.
Annotation Using Syglass
Alternative to Napari, you may use Syglass to annotate the volume. The steps are as follows:
Acquire license key and prepare VR equipments and a VR platform such as SteamVR
Open Syglass and create a new project from the ome zarr image, then right click the project to “Add Mask” to the volume
In the Syglass, click the VR button to turn on VR, connecting to SteamVR, at which point you should see headset display the Syglass environment
Open the project, and adjust the settings in the menu to optimize data display; use the ROI tool to annotate the volume
Click the save button in ROI tool menu to save annotation; the saved ROI can be exported as a stack of tiff files using the Project > ROI tab
Misc:
ROI tool has an important setting named resolution level; when painting, set this to max since we need the highest mask resolution possible to accurately capture the edges of the brain region. Also turn on the fixed resolution level setting just below it.
Cross sectioning tool has the function to display raw data patch, but this 2d view uses the same set of settings as 3d view which often would not be able to optimize both. But you can save a separate setting of threshold, window, brightness… etc. by using the “1” “2” “3” buttons located beside the save icon in the settings menu.
The engineers at Syglass is very helpful and responsive to requests, contacted at “info@syglass.io”. I have solved a few issues with OME ZARR and Syglass usage by contacting support this way.